An Important Step Regarding First Human Migration Routes and Disease Related Genes
The first results of an international project on the genomic landscape of the Mediterranean Basin and the Middle East, which also included Turkish scientists and universities, were published in the September 2016 issue of the journal, Nature Genetics.
Humans started migrating to other parts of the world from Africa about 100,000 year ago. Small groups first reached the Arabian Peninsula and Mesopotamia, and then the continents of Asia, Australia, Europe and America. Details of these journeys have been tracked for many years by examining fossils and Stone Age artifacts, and through comparative linguistics. The science of genetics has become one the most important components of archeological studies, following groundbreaking conceptual developments on biological roots of inheritance, from the 19th century onwards and particularly with the development of recombinant DNA technology in the second half of the 20th century. From these developments, a hybrid scientific discipline called archaeogenetics has emerged.
The world of science had been waiting impatiently for a map of the genomic landscape of the Mediterranean basin and the Middle East, from where the first civilizations in human history emerged and about 10 percent of the world’s population currently lives. The first results, shared with the world scientific community in the September 2016 issue of the journal Nature Genetics, convey two important observations. First, the high rates of consanguinity observed in these societies speed up the process of discovering disease related genes by a factor of 4 to 7. Secondly, mapping the genomic landscape of this large geographical region sheds light on identifying the migration routes to Asia and Europe.
The project, which grew out of publications by an international team that met in Ankara in 2009 and 2010 and was led by Prof. Dr. Tayfun Özçelik, member of TÜBA’s (Turkish Academy of Sciences) Academy Council and a faculty member at Bilkent University, was carried out over five years with a project budget of tens of millions of dollars, with the participation of 180 researchers from Turkey, Egypt, Israel, Syria, Iraq, Jordan, Lebanon, Saudi Arabia, United Arab Emirates, Qatar, Iran, Pakistan, Morocco, Algeria, Libya and Tunisia, and tens of universities including Rockefeller, Harvard, MIT, California, Cornell, INSERM, Alfaisal and Istanbul. The project’s findings were presented to the global scientific community in a lead article by Prof. Dr. Tayfun Özçelik and Dr. Onur Emre Onat.
As part of the project, initially, a DNA bank was created with the support of the Greater Middle East Variome Consortium, which includes 30 scientists from Turkey. Following this, genomes of 1,111 volunteers were examined using next generation sequencing.
Migration Routes and the History of Humanity
In the first stage of the study, comparisons were carried out with the 1000 Genomes project, which examined European, Asian, African, Australian and American societies. Significant differences were found within Mediterranean societies from Northwest Africa to Southeast Asia, and it was found that some regions were grouped together with European and Asian societies. Routes of migration, which took place over a period of about 100,000 years, and which during their first stage led from Africa to Mesopotamia and Anatolia, and through the Arabian Peninsula, Iran, Pakistan, Indian coasts and Oceania islands to Australia, were examined using the genome sequences of people currently living in these regions. Valuable information was obtained about the genetic diversity of people who migrated to Europe over Central Asia and Anatolia. The region which covers the modern-day provinces of Urfa, Mardin and Gaziantep, and which had human settlements prior to Göbekli Tepe, is a of key importance. Genetic distances between societies inhabiting the Northwest African coast, Arabian Peninsula, Iran and Pakistan were found to be large, similar to the distance between the Finns and Tuscanians (in Italy), and after Turkey, Syria was the country that had the biggest similarity with European societies. In terms of genetic diversity, Turkey had the highest values. The central role played by Anatolia in the migration of humans to Europe, which was previously explained on the basis of archaeological, linguistic, cultural and historical findings, was thus established at a genomic level.
Routes of migration out of Africa
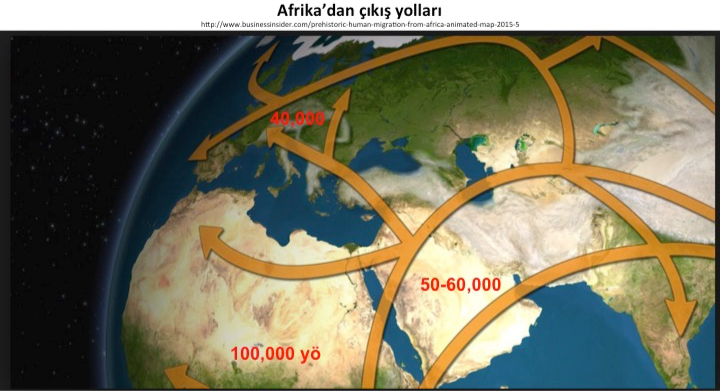 |
Routes of migration into other parts of the world
.png) |
Migration of Modern Humans to Europe
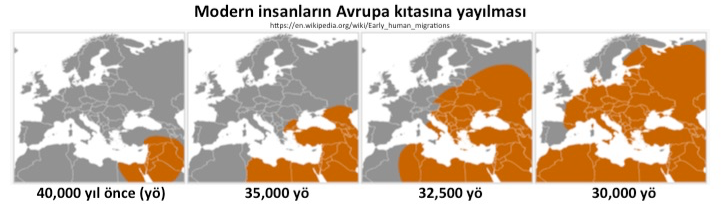 |
Finding Disease-Related Genes
Genome projects, particularly those involving the human genome, have become very prominent over the last two decades. Identifying disease related genes and developing early diagnosis tests and molecular targeted treatment options have become one of the most important priorities of modern medicine. At this point, the family structures of different societies play an important role. Consanguinity and having multiple children facilitate the interpretation of genomes. Homozygosity mapping over chromosomes has shown that sizes of four megabases and above were obtained only in Anatolian, Mesopotamian, Northwest African and Southeast Asian societies.
This presents a unique opportunity to interpret high-technology based genome data and offering it to the service of humanity. Indeed, it was found that the chances of discovering a gene related to a group of neurogenetic disorders that interfere with brain development are 4 to 7 times higher when compared with America, Europe and China, and the chances of success increase by 700 percent.
A New Hypothesis
Genomic sciences have achieved great success over the last two decades in identifying the genes for rare diseases, such as cystic fibrosis, phenylketonuria, ataxia and developmental disorders, which are also known as single gene disorders and number more than 5000. It should be emphasized that, even though these diseases are rare individually, together they make up the largest group of diseases, especially in childhood. However, a similar success has not yet been recorded in identifying genes for obesity, movement disorders, sleep problems, migraine, high blood pressure, diabetes and some cancers, which are more widespread among adult populations, and disproportionately affect aging societies. The most important hurdle is a conceptual one. In the leading article he co-authored with his student, Dr. Emre Onat, in Nature Genetics, Dr. Özçelik presented to the scientific community a new conceptual approach and roadmap that can be used to examine the human genome. The article argues that this approach, which was used in the identification of the genes for Uner Tan Syndrome -defined first in Hatay in 2000s, characterized by quadrupedal gait, and named after an honorary TÜBA member - and later in the identification of genes for Parkinson’s and athetosis diseases, can be used to examine the genetic roots of many complex diseases.
40,000 years ago 35,000 years ago 32,500 years ago 30,000 years ago
 |
Also known as precision medicine, this new genome project, which started in 2015 by the joint initiative of the USA, EU and China, supported by the Tokyo statement this year by science academies, including TÜBA, of G20 countries, and expected to result in a fundamental change in the diagnosis and treatment of diseases by 2030, involves achieving a molecular understanding of complex diseases, using new conceptual approaches, and is among the most important priorities of humanity today

